Research
Prof. Rafail Ostrovsky & Prof. Sam Kumar (Multi-party Computation)
Nov 2023 - Current
Under the guidance of Professor Rafail Ostrovsky and Professor Sam Kumar, I am deeply engaged in the development of a pioneering data collaboration service leveraging multi-party computation (MPC) technologies, including MP-SPDZ and EMP Toolkit. This groundbreaking initiative is aimed at enabling the secure exchange and processing of data within an MPC framework on AWS EC2 virtual machines.
At the heart of our project is the design of an automated system that streamlines the setup of AWS EC2 instances and ensures the secure transfer of input data via MPC. A key feature of our system is the establishment of a TLS connection with AWS, which facilitates the secure execution of computations on the cloud platform. Our approach involves a detailed analysis of web requests to AWS, translating them into the MPC framework's domain-specific language (DSL), thus bolstering data privacy through advanced encryption and secure secret sharing techniques inherent to MPC.
Looking ahead, we plan to enrich the system with state management features, which will enable the service to monitor and restore its state, thereby enhancing its resilience and introducing reactive capabilities that will significantly improve the service's efficiency and user experience.
Our vision is to transform MPC into a versatile platform that functions like an API, enabling secure data collaboration in a manner akin to Snowflake's data cleanroom, thereby redefining the possibilities of secure data interaction in the cloud.
Prof. Ostrovsky Website Prof. Kumar WebsitePimentel Lab UCLA Health
Sep 2022 - Current
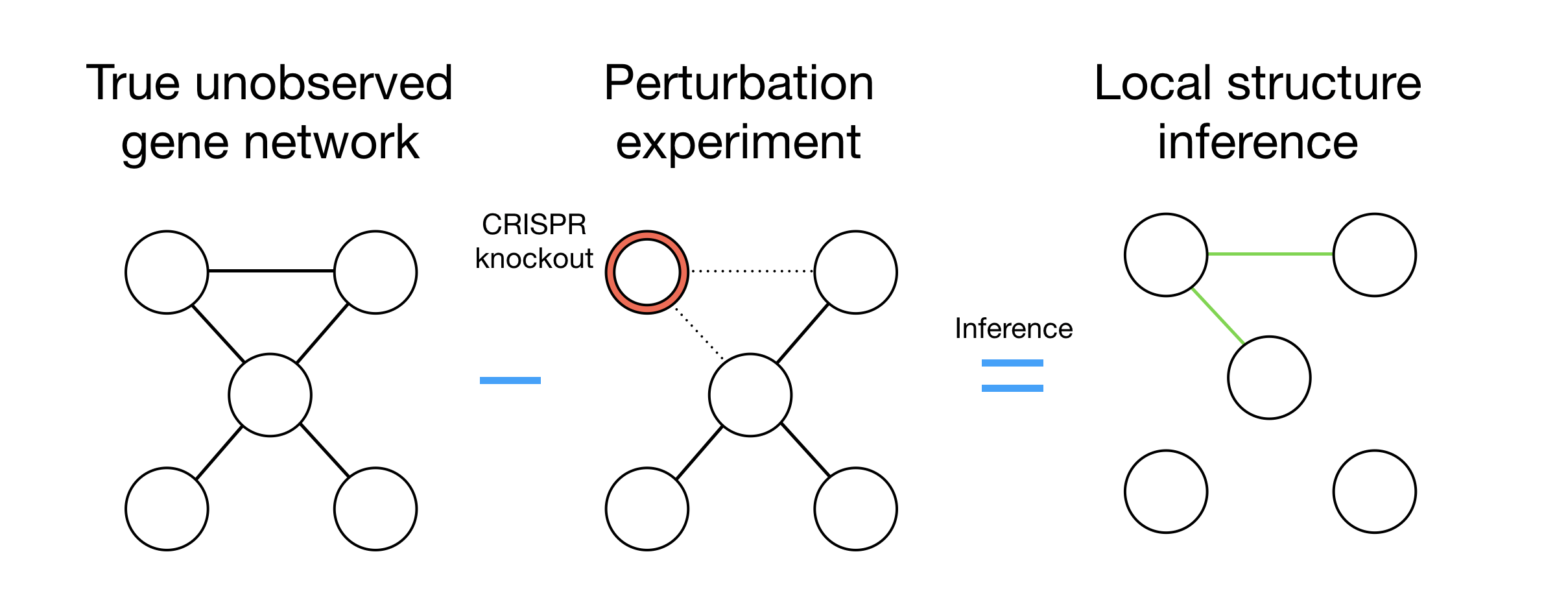
At the Pimentel lab, I was deeply engaged in advancing our understanding of gene regulation and the impact of diseases on gene networks. My focus was on developing and optimizing an automated pipeline that significantly enhanced our data processing capabilities. By automating the retrieval and processing of 4,865 SRA files, employing Kallisto for precise read alignment, and utilizing Snakemake along with CLI scripting for data normalization, I managed to save the lab an invaluable amount of time—equivalent to over two weeks of manual labor.
Further, I applied my knowledge in Bayesian statistics and structural learning, utilizing Python's robust libraries like Numpy, Scipy, Matplotlib, and Seaborn, to process and analyze genome wide perturb seq data. Through meticulous data analysis, I identified anomalies within the dataset—instances where the data failed to meet the expected threshold z-scores. This critical insight led to the saving of more than 100 lab hours, optimizing our research efficiency and contributing to more accurate and reliable outcomes in our studies of gene regulation networks.
Lab Website